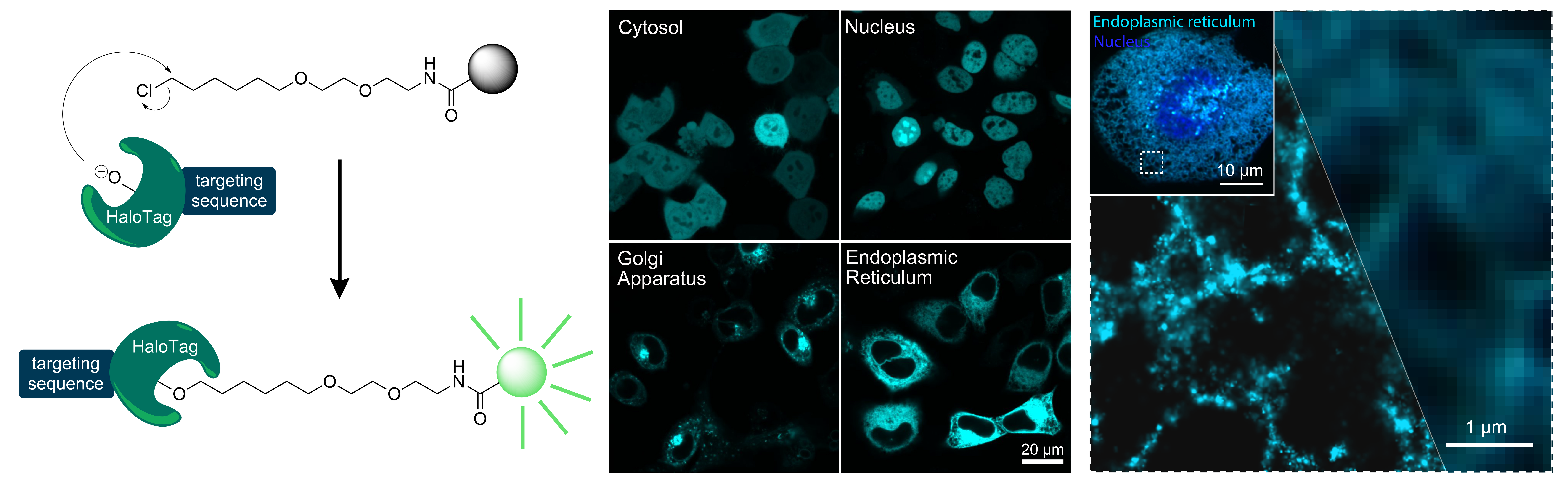
Development of dyes and labeling strategies for monitoring physiological processes in eukaryotic and prokaryotic cells
Fluorescence microscopy is an invaluable tool to observe live biological samples, but its resolution is limited to about 200 nm by diffraction of visible light. Single-molecule localization microscopy techniques surpass this limit by using photoactivatable or spontaneously blinking fluorophores. In our group, we design and synthesize novel fluorescent probes that offer control of fluorescence emission by photoactivation, enzymatic activation, spontaneous thermal switching, response to the physicochemical environment, or a combination of these. To provide a highly specific fluorescence signal and a universally applicable labeling strategy for different molecular targets, we utilize self-labeling proteins, e.g. HaloTag and SNAPtag.
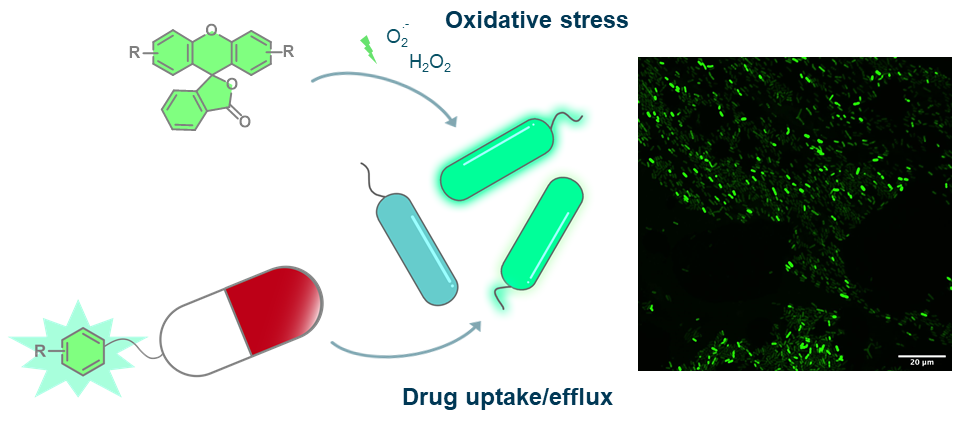
The emergence of bacteria with resistance against antibiotics is one of the most serious threats that we face. To develop novel strategies to fight resistant pathogens, we have to understand the physiological properties of bacteria not only in laboratory culture, but within human patients. As part of the NCCR AntiResist, we design and synthesize xanthene-based probes that utilize a combination of chemical reactions and bacterial enzymatic activity to investigate oxidative stress in bacterial samples from human infections. Furthermore, we modify established antibiotics to monitor drug uptake and efflux in these bacterial clinical samples.
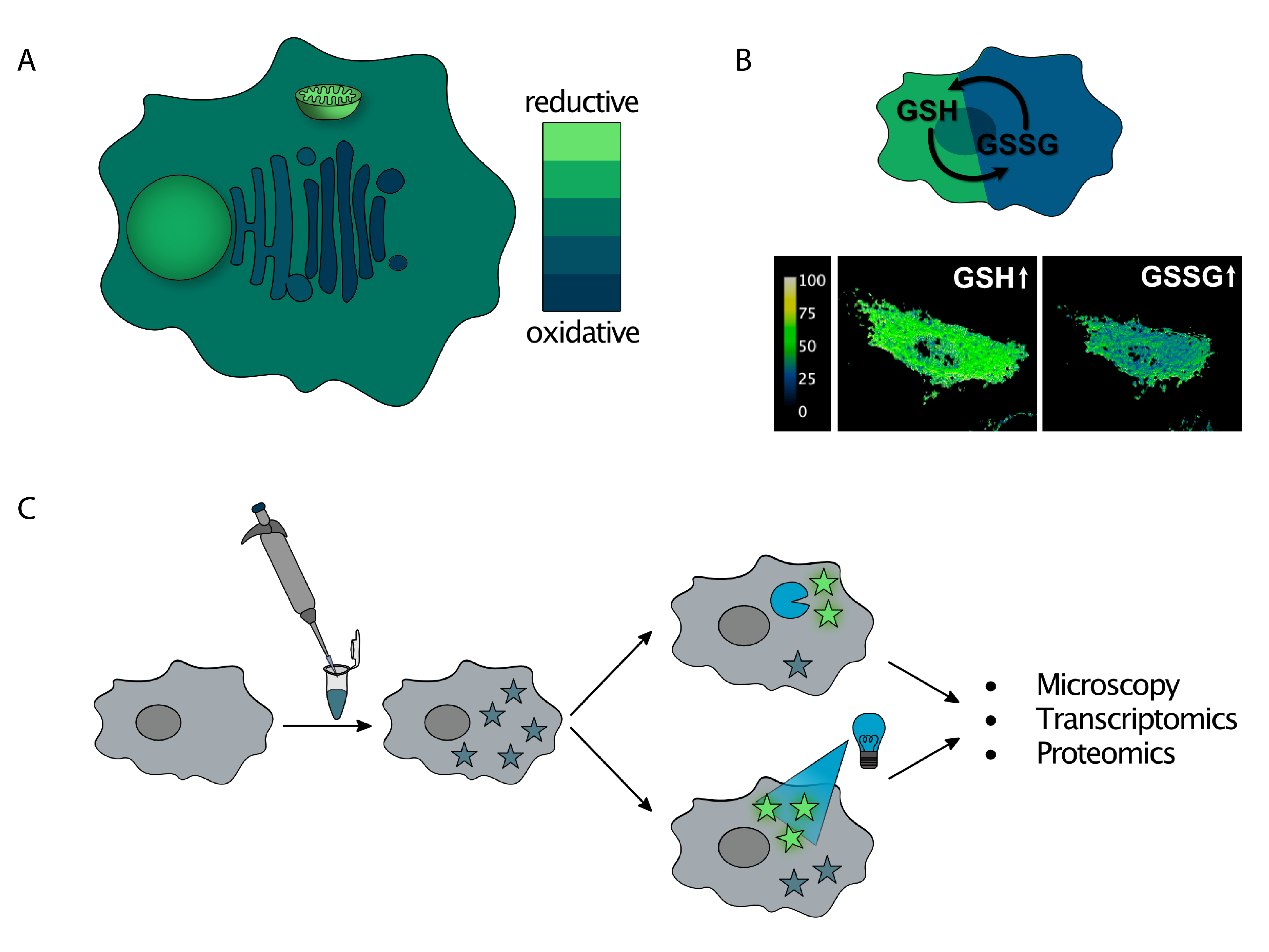
Characterization and manipulation of redox homeostasis in subcellular compartments
Redox homeostasis is essential for cell function and plays a role in metabolic and cell signaling pathways. Our group focuses on mapping the redox state across the different comparmtents of the cell (A). We develop molecular and biological probes that can monitor levels of reduced and oxidized glutathione (GSH/GSSG), the main redox buffer of the cell (B). Moreover, we study how the cell responds to redox imbalances such as reductive stress. For this purpose, we use chemical probes that undergo photolytic or enzymatic activation, generating a reducing agent and a fluorophore. After the stress is locally generated, organellar stress-response pathways can be studied using a combination of microscopy techniques and omics analyses (C).
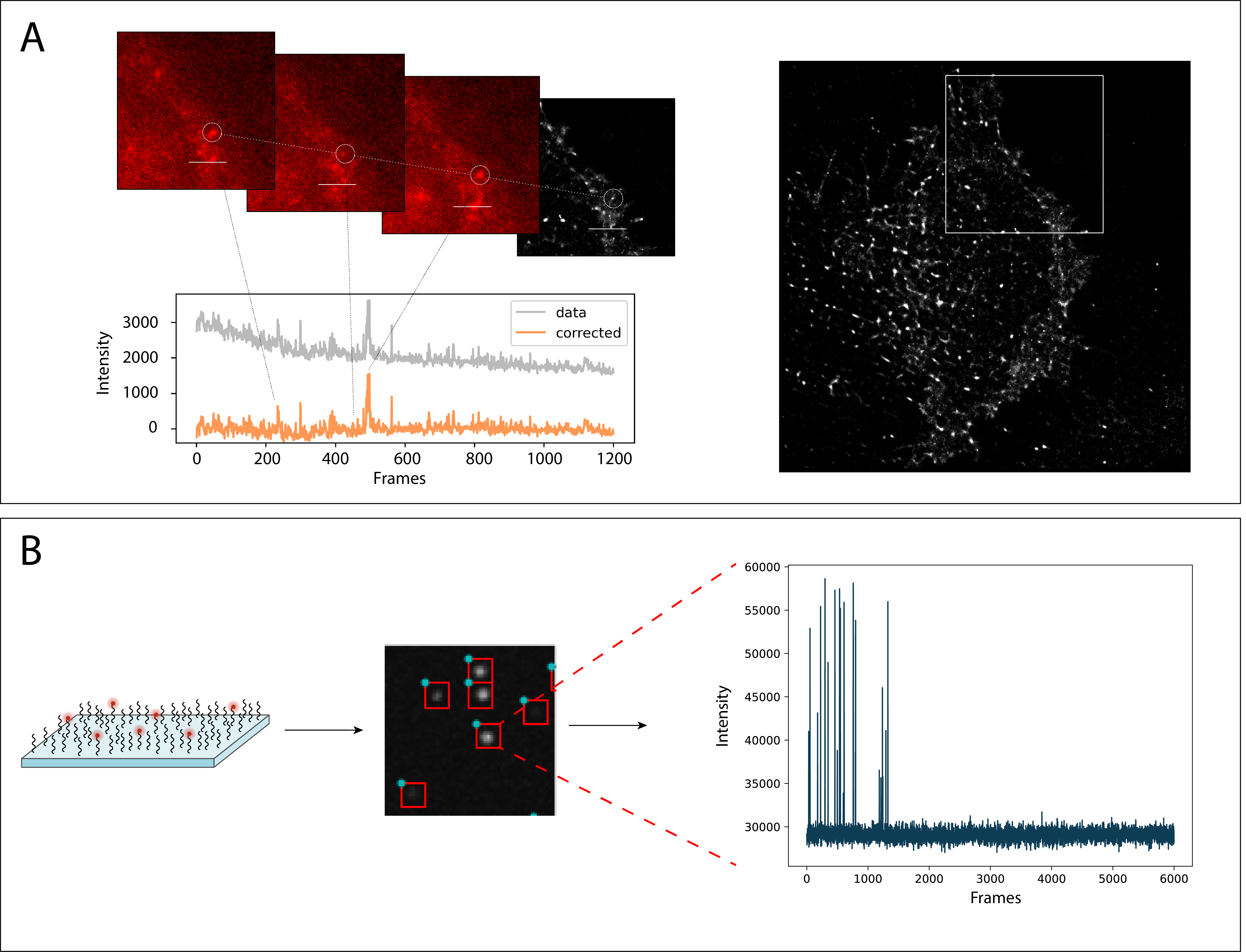
Single-molecule imaging in vitro and in live cells
Photoactivatable or spontaneously thermally blinking fluorophores allow for high-resolution imaging using single-molecule localization microscopy (SMLM). One focus of our research is on combining spontaneously blinking fluorophores with methods like FRET (A) and surface conjugation chemistry (B). Together with computational image analysis and machine learning, we can gain detailed information of biomolecular structure and interactions.